Linear Regression: Jupyter Notebook#
pandas.DataFrame#
Reading Data#
Using read_csv() from pandas, read data into dataframe. If your data happens to be in a M$ Excel file, then there is also a read_excel() function.
[1]:
import pandas as pd
[2]:
dataset = pd.read_csv('./history_data.csv')
Relationship Between pandas.DataFrame and numpy.ndarray#
See how a DataFrame holds values using numpy.ndarray.
[3]:
dataset.values
[3]:
array([['New York', 'New York', nan, ..., 0.0, nan, 'Clear'],
['New York', 'New York', nan, ..., 0.0, nan, 'Clear'],
['New York', 'New York', nan, ..., 0.0, 87.77, 'Clear'],
...,
['New York', 'New York', nan, ..., 23.3, 74.96, 'Clear'],
['New York', 'New York', nan, ..., 14.3, 70.33, 'Clear'],
['New York', 'New York', nan, ..., 0.0, 84.26, 'Clear']],
dtype=object)
[4]:
type(dataset.values)
[4]:
numpy.ndarray
For convenience, pandas.DataFrame provides many attributes from the underlying numpy.ndarray.
Two dimensional array …
[5]:
dataset.ndim
[5]:
2
… extending such and such cell in each direction …
[6]:
dataset.values.shape
[6]:
(72, 16)
[7]:
dataset.shape
[7]:
(72, 16)
DataFrame.describe() is convenient for interactive use in a Jupyter notebook, just like many other methods.
[8]:
dataset.describe()
[8]:
| Resolved Address | Maximum Temperature | Minimum Temperature | Temperature | Wind Chill | Heat Index | Precipitation | Snow Depth | Wind Speed | Wind Gust | Cloud Cover | Relative Humidity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 0.0 | 72.000000 | 72.000000 | 72.000000 | 0.0 | 22.000000 | 72.000000 | 0.0 | 72.000000 | 65.000000 | 72.000000 | 38.000000 |
| mean | NaN | 82.995833 | 70.688889 | 76.287500 | NaN | 91.327273 | 0.012222 | NaN | 12.690278 | 20.470769 | 3.369444 | 72.513421 |
| std | NaN | 5.946106 | 5.574023 | 5.313256 | NaN | 5.994536 | 0.070695 | NaN | 6.025980 | 8.302423 | 7.214825 | 12.665492 |
| min | NaN | 69.900000 | 53.300000 | 61.500000 | NaN | 82.400000 | 0.000000 | NaN | 2.200000 | 4.700000 | 0.000000 | 46.090000 |
| 25% | NaN | 79.050000 | 68.900000 | 74.300000 | NaN | 87.050000 | 0.000000 | NaN | 9.100000 | 15.000000 | 0.000000 | 66.377500 |
| 50% | NaN | 83.750000 | 71.950000 | 76.650000 | NaN | 89.750000 | 0.000000 | NaN | 12.800000 | 19.700000 | 0.000000 | 72.330000 |
| 75% | NaN | 87.950000 | 74.325000 | 79.850000 | NaN | 95.950000 | 0.000000 | NaN | 15.425000 | 25.300000 | 2.825000 | 81.715000 |
| max | NaN | 92.900000 | 80.700000 | 85.800000 | NaN | 101.600000 | 0.470000 | NaN | 38.000000 | 50.600000 | 34.500000 | 96.970000 |
Extracting Input and Output Features from a pandas.DataFrame#
Beware that algorithms expect a two dimensional array as the set of inputs. Using the column header (“Minimum Temperature”) to index the dataframe gives a list-like type. Wrong!!
[9]:
inputfeatures = dataset['Minimum Temperature']
[10]:
inputfeatures
[10]:
0 53.3
1 58.7
2 60.2
3 66.8
4 68.3
...
67 70.1
68 72.2
69 72.1
70 75.5
71 78.2
Name: Minimum Temperature, Length: 72, dtype: float64
[11]:
type(inputfeatures)
[11]:
pandas.core.series.Series
Correction: index the dataframe with a list of column headers
[12]:
inputfeatures = dataset[['Minimum Temperature']]
Sign of correctness: that one is made up more nicely by the notebook:
[13]:
inputfeatures
[13]:
| Minimum Temperature | |
|---|---|
| 0 | 53.3 |
| 1 | 58.7 |
| 2 | 60.2 |
| 3 | 66.8 |
| 4 | 68.3 |
| ... | ... |
| 67 | 70.1 |
| 68 | 72.2 |
| 69 | 72.1 |
| 70 | 75.5 |
| 71 | 78.2 |
72 rows × 1 columns
Likewise, the output features.
[14]:
outputfeatures = dataset[['Maximum Temperature']]
Plotting with matplotlib#
Fortunately (well, that was on purpose), our feature sets are one dimensional, so plotting the dataset im two dimensions makes sense. Multidimensional data analysis is not so straightforward - this is why they call it data science.
[15]:
import matplotlib.pyplot as plt
pandas.DataFrame interacts nicely with matplotlib.
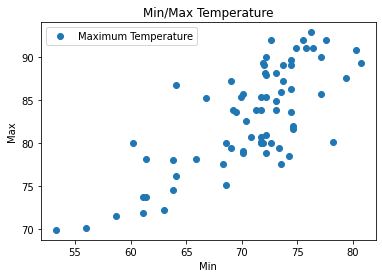
[16]:
dataset.plot(x='Minimum Temperature', y='Maximum Temperature', style='o')
plt.title('Min/Max Temperature')
plt.xlabel('Min')
plt.ylabel('Max')
plt.show()
Data Splicing: Split into Training and Test Data#
Before creating the model (from an algorithm and a dataset), we prepare the dataset
80% for training
20% for testing/verification
[17]:
import sklearn
from sklearn.model_selection import train_test_split
[18]:
input_train, input_test, output_train, output_test = \
train_test_split(inputfeatures, outputfeatures, test_size=0.2, random_state=0)
Creating the Model: Algorithm + Training Data#
[19]:
from sklearn.linear_model import LinearRegression
Initially, the model is the algorithm
[20]:
model = LinearRegression()
Next, we feed it the training data
[21]:
model = model.fit(input_train, output_train)
Model is complete; see the parameters of the linear interpolation (would need theory to better understand):
[22]:
model.coef_
[22]:
array([[0.80189231]])
[23]:
model.intercept_
[23]:
array([25.95355086])
Verify the Model#
We saved 20% of the dataset for verification.
Use the model to predict the output for the input test data.
Compare prediction to actual output test set
[24]:
output_predicted = model.predict(input_test)
Here we (ab)use a pandas.DataFrame to nicely format actual output test data and predicted output side by side.
Note that input_test is a pd.DataFrame, but output_predicted is a numpy.ndarray.
Reason:model.predict() is happy with anything that supports indexing (thanks to duck typing - we gave it a Dataframe), but its output is always a numpy.ndarray
[25]:
pd.DataFrame({'Actual': output_test.values.reshape((15,)),
'Predicted': output_predicted.reshape((15,))})
[25]:
| Actual | Predicted | |
|---|---|---|
| 0 | 80.0 | 83.609608 |
| 1 | 84.9 | 84.571879 |
| 2 | 91.1 | 86.736988 |
| 3 | 80.0 | 84.170933 |
| 4 | 78.2 | 78.798254 |
| 5 | 92.0 | 84.170933 |
| 6 | 78.2 | 75.189739 |
| 7 | 92.0 | 88.180394 |
| 8 | 85.4 | 83.449230 |
| 9 | 80.1 | 88.661530 |
| 10 | 92.9 | 87.057745 |
| 11 | 85.4 | 83.850176 |
| 12 | 87.2 | 81.284120 |
| 13 | 90.0 | 83.850176 |
| 14 | 83.6 | 81.685067 |
Comparing the actual and predicted values, we can see that they are “not far off”. Whatever this means - in a real data science world (this is only the surface), we would now have to use advanced statistical methods to actually measure the term “not far off”.
But this is left to data scientists. Our job is to create correct programs, and to keep those maintainable.
